Systemic Lupus Erythematosus (SLE) is an autoimmune disease with diverse manifestations, from mild and sporadic to severe and life-threatening. SLE heterogeneity arises from various factors, including genetic, epigenetic, and immune niche-related features (Kaul et al., 2016; Jenks et al., 2018). However, the exact interactions and hierarchy between these features remain unclear. Recent studies have found somatic mutations in genes typically mutated in B-cell lymphomas, in subsets of B-cells that are associated with SLE pathogenesis (Piotrowski et al., 2015; Pullabhatla et al., 2018; Singh et al., 2020; Brown et al., 2022). These mutations may support B-cell clonal bursts, but direct information from clinical specimens on the impact of these mutations on B-cell lineages and their influence on SLE progression or malignant transformation is missing.
To define the somatic mutational landscape of B-cells in SLE, we first profiled peripheral blood mononuclear cells obtained from a pilot cohort of four SLE patients using whole-exome sequencing (WES; 200X depth). Following GATK best practices (Van der Auwera and O'Connor, 2020), we identified somatic alterations in DDX11 [variant allele frequency (VAF): 10.2%], DNMT3A [VAF: 3.4%]; PMS2 [VAF: 47.9%], and BICD1 [VAF: 47.3%]; these genes have been linked to lymphoid malignancies and SLE pathogenesis (Piotrowski et al., 2015; Pullabhatla et al., 2018; Niroula et al., 2021; Saeed et al., 2021). We validated the presence of the top-identified mutations using droplet digital PCR technology on sorted B-cell subsets (naïve, memory, and double-negative [DN; CD19 hiIgD loCD27 lo]) from these SLE patients. DN cells are a heterogeneous subtype of autoreactive memory B-cells that play a crucial role in SLE pathogenesis being the precursors of autoantibody producing plasma cells (Wei et al., 2007; Jenks et al., 2018).
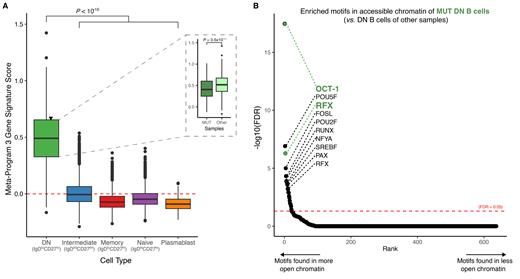
To investigate the transcriptional and epigenetic alterations in the mutation-harboring B-cells, we then conducted single-cell multi-omic sequencing. We profiled sorted activated B-cells (CD19 hiIgD lo) from the four SLE cases, to largely exclude non-autoreactive naïve B-cells. To maintain a comparison with naïve B cells, we included a small proportion of these cells in the input. In total, we analyzed 35,339 cells, and identified different B-cell subsets with distinct gene expression patterns, including DN cells ( n = 1,437 cells; ZEB2 hiTRAF5 lo, Jenks et al., 2018). To uncover, in an unbiased way, transcriptional differences between B-cell subsets, we catalogued variation in transcriptomic signatures derived from the scRNA data using consensus non-negative matrix factorization. We found a unique gene expression program exclusively present in DN cells ( Panel A), which includes key genes implicated in immune regulation and the development of autoimmunity, such as PTPN22 (Chung & Criswell, 2007; Tizaoui et al., 2021) . Pathway analysis of the DN-specific program revealed an enrichment of complement system genes (FDR = 0.074; Hallmark), which are crucial for immune regulation and tolerance, such as CBLB (Tang et al., 2019). Notably, in DN cells from one of the SLE samples carrying lymphoma/SLE-associated mutations in BICD1, PMS2, DDX11, we observed an altered expression of this gene program compared to DN cells from other samples ( P-value = 3.6x10 -11; Panel A, inset), suggesting that a malfunctioning complement system in DN cells may lead to an overly active immune response (Weinstein et al., 2021). In line with this notion, using chromatin accessibility profiles as a measure of transcription factor (TF) regulatory activities, we showed increased activity of TFs involved in interferon-dependent innate immune response and in B-cell activation and differentiation (RFX and OCT-1, respectively; FDR < 0.05), in the DN B-cells of this sample ( Panel B).
In summary, we uncovered a unique transcriptional program associated with autoimmune DN B-cells. This program showed enrichment in key genes and pathways involved in immune response regulation. The disruption of this program, accompanied by the somatic SLE-associated mutations, may lead to overactivation of DN B cells, ultimately contributing to autoimmunity. Further analyses involving a larger cohort of clinical specimens are needed to directly link B-cell genotypes with their transcriptional and epigenetic programs and to elucidate the role of the identified mutations in SLE pathobiology.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal